“SARS-CoV-2 D614G variant enhances infectivity, replication and transmission - News-Medical.Net” plus 3 more
“SARS-CoV-2 D614G variant enhances infectivity, replication and transmission - News-Medical.Net” plus 3 more |
- SARS-CoV-2 D614G variant enhances infectivity, replication and transmission - News-Medical.Net
- Biopharma Update on the Novel Coronavirus: September 30 - BioSpace
- CRISPR screening identifies human host pathways that facilitate coronavirus infection - News-Medical.Net
- In deadly COVID-19 lung inflammation, discover a culprit in NFkB pathway - Science Daily
| SARS-CoV-2 D614G variant enhances infectivity, replication and transmission - News-Medical.Net Posted: 30 Sep 2020 04:09 PM PDT Researchers in the United States and Japan have conducted a study showing a common mutation in the spike protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) - the agent that causes coronavirus disease 2019 (COVD-19) - enhances the infectivity, replication, and early transmission of the virus.  The team's study of SARS-CoV-2 engineered to harbor the D614G mutation found that this strain was replicated more efficiently in primary human proximal airway epithelial cells than the wildtype virus did. In a hamster model of infection, the D614G strain also showed much faster respiratory droplet transmissibility than the wildtype virus shortly following infection. Ralph Baric (the University of North Carolina at Chapel Hill) and colleagues from the University of Wisconsin, University of Tokyo, and the National Institute of Infectious Diseases, Tokyo, say the findings support the need to periodically review SARS-CoV-2 contemporary isolates and identify any new variants with increased transmissibility and pathogenesis that may have emerged. A pre-print version of the paper is available on the server bioRxiv*, while the article undergoes peer review. 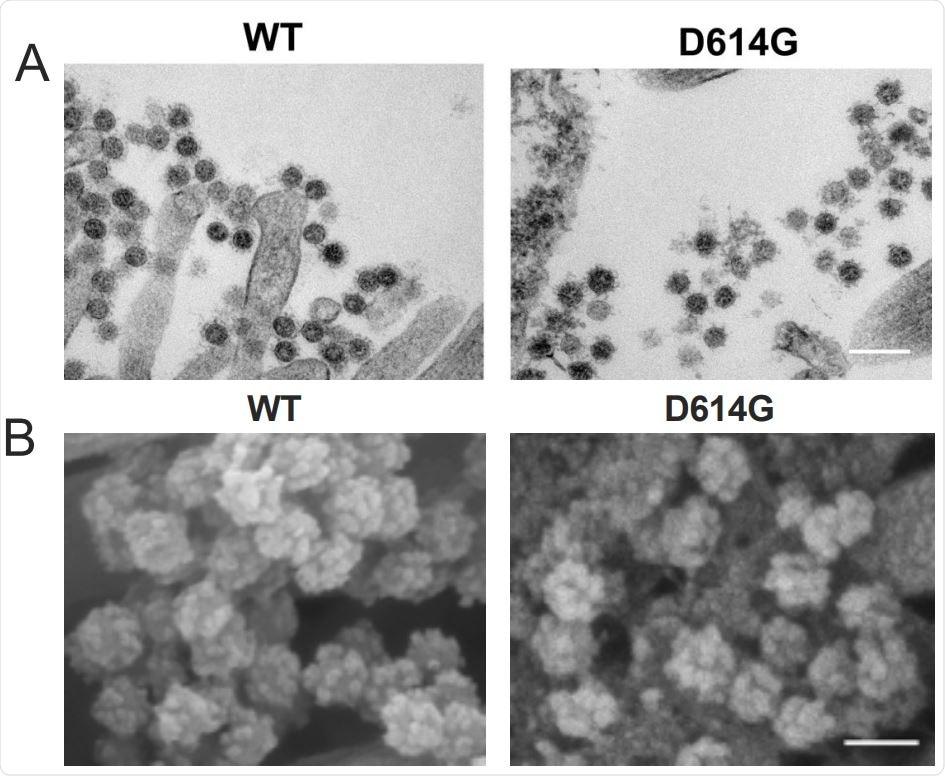 D614G substitution does not alter SARS-CoV-2 virion morphology and S protein cleavage pattern but change viral sensitivity to neutralizing antibodies. A. Transmission electron microscopy image of WT and D614G virions on airway epithelial cell surface, scale bar: 200 nm. B. Scanning electron microscopy images of WT and D614G virions on airway epithelial cell surface, scale bar: 100 nm. The importance of the spike proteinSince the first cases of SARS-CoV-2 infection were identified in Wuhan, China, late last year, the virus has continued to sweep the globe and has now infected more than 33.8 million people and caused more than one million deaths. Although most people who become infected only develop mild or asymptomatic disease, some develop severe health complications such as cardiac problems, coagulopathy, stroke, or acute respiratory distress syndrome. To gain entry to host cells, SARS-CoV-2 uses a surface structure called the spike glycoprotein to bind the human cellular receptor angiotensin-converting enzyme 2 (ACE2). This spike protein has, therefore, become a central focus of interest in studies aiming to develop vaccines and therapies. During its pandemic spread in naïve populations, a virus may select for mutations that change its virulence, pathogenesis, or transmissibility. Studies have recently identified the D614G substitution in the spike glycoprotein as the most prevalent strain of SARS-CoV-2 circulating globally. However, the effects of this variant on the function, pathogenesis, and transmissibility of SARS-CoV-2 remain unclear. What did the researchers do?To investigate the function of the D614G substitution in SARS-CoV-2 replication and transmissibility, the researchers engineered variants containing the D614G mutation in the spike protein, as well as a second variant containing the gene for the bioluminescent reporter nanoLuciferease (nLuc). The team compared the growth of wildtype SARS-CoV-2 and the D614G variant in primary human nasal epithelia (HNE), large (proximal) airway epithelia (LAE), and distal lung small airway epithelia (SAE). The D614G-infected HNE and LAE cultures, but not the SAE cultures, exhibited significantly higher viral titers than the wildtype-infected cultures. Competitive co-infection assays performed in LAE cultures simultaneously infected with both viruses showed that the D614G variant became dominant in the cultures, whether the wildtype virus was originally present at a 1:1 or 10:1 ratio over the D614G mutant.
Next, the team performed scanning and transmission electron microscopy to visualize virions present on the surface of primary human airway cell cultures. No significant differences in virion morphology or the number of spike proteins were observed between the two viruses. Further analysis revealed more differences between the virusesThe researchers used the nLuc-expressing recombinant SARS-CoV-2 encoding either wildtype or D614G spike to measure antibody neutralization activity in serum samples taken from mice vaccinated with D614 (wildtype) spike. This revealed that the samples half-maximal inhibitory dilution values against the D614G virus were between 0.8 and 5.1 times higher than against the wildtype virus, indicating that the D614G variant makes SARS-CoV-2 more sensitive to neutralizing antibodies. Evaluating respiratory droplet transmissibilityTo evaluate the role of the D614G variant in SARS-CoV-2 respiratory droplet transmissibility, the researchers set up eight pairs of hamsters, each comprising a naïve hamster alongside an infected animal 1 day following infection. Both the wildtype and D614G viruses were efficiently transmitted to naïve hamsters. At 4 and 6 days following infection, the infected hamsters and the exposed hamsters exhibited similar viral titers, regardless of which virus they had been infected with. However, five of eight hamsters exposed to the D614G-infected group showed infection and had detectable viral shedding on day 2, while those exposed to the wildtype-infected group showed no infection or viral shedding. This suggests that the D614G variant is transmitted much more quickly between hamsters via droplets and aerosols than the wild type virus is.
*Important NoticebioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information. Journal reference: |
| Biopharma Update on the Novel Coronavirus: September 30 - BioSpace Posted: 30 Sep 2020 08:58 AM PDT News information is not all-inclusive and updates are published on Mondays, Wednesdays and Fridays. FDA Actions The U.S. Food and Drug Administration (FDA) accepted Windtree Therapeutics' Investigational New Drug (IND) application for a Phase II trial for its KL4 surfactant drug. The drug will be tested in COVID-19-associated lung injury and ARDS patients. Diagnostics President Donald Trump announced that 150 million units of Abbott's BinaxNOW COVID-19 Ag Card Point of Care (POC) SARS-CoV-2 diagnostic test will be distributed across the country. Abbott's test was designed to deliver results within 15 minutes without the necessity of a laboratory. Becton, Dickinson and Company received a CE mark for its rapid, point-of-care, SARS-CoV-2 antigen test for use on the BD Veritor Plus System. The test delivers results in 15 minutes. The company expects commercial availability of this new assay at the end of October for countries in Europe that recognize the CE mark. Avacta Group announced its BAMs assay to detect COVID-19 is now in use as a research kit, courtesy of partner company Adeptrix. The bead-assisted mass spectrometry (BAMS) assay uses the Affimer reagents specific to the SARS-CoV-2 virus to capture the virus spike protein from the sample for rapid detection by mass spectrometry. Up to one thousand samples per day can be analyzed by a single technician using BAMS. Testing Therapies, Antivirals and Vaccines Russia announced it will share preliminary data from the first six weeks of the ongoing Phase III study for its already-approved COVID-19 vaccine. Russia's vaccine is an adenoviral vector vaccine developed by Moscow's Gamaleya Institute. Regeneron Pharmaceuticals announced early data from its Phase I/II/III trial of its antibody cocktail, REGN-COV2, against COVID-19. The analysis showed that the antibody cocktail decreased viral load and alleviated symptoms faster in non-hospitalized COVID-19 patients. Windtree Therapeutics, Inc. will initiate Phase II studies of lyo lucinactant, a KL4 surfactant drug, in COVID-19 associated lung injury and acute respiratory distress syndrome (ARDS) patients. Algernon Pharmaceuticals has enrolled 100 patients, two-thirds of its target, in a Phase II/III study of NP-120 (Ifenprodil), an N-methyl-D-aspartate (NMDA) receptor antagonist, for the treatment of COVID-19. The study is expected to be completed in November with a planned data readout before the end of the fourth quarter. Moderna published second interim analysis of its open-label Phase I trial of its COVID-19 vaccine, mRNA-1273. This particular data focused on two age cohorts, one ages 56-70 and the other 71+. Company Actions/Announcements Grand River Aseptic Manufacturing (GRAM) inked a deal with Janssen Pharmaceutical, a Johnson & Johnson company, to support the manufacturing of Janssen's COVID-19 vaccine candidate. The agreement with GRAM includes technical transfer and fill-and-finish manufacture of the vaccine candidate. The FDA granted expanded access to Organicell Regenerative Medicine for its proprietary therapeutic, Zofin as a treatment of COVID-19. This expanded access protocol will provide access to the investigational product Zofin for patients who have mild to moderate COVID-19, or who are judged by a healthcare provider to be at high risk of progression to moderate disease. Other Industry News According to new research published in the Journal of Fluid Mechanics, ventilation systems in many modern office buildings may increase the risk of coronavirus exposure, especially in the coming winter months. Researchers found that the ventilation systems disperse airborne contaminants, which could potentially include droplets and aerosols containing viruses. |
| Posted: 27 Sep 2020 08:35 PM PDT The human coronavirus family consists of 7 known pathogens, for which there are no approved vaccines or specific therapeutic options. Although the seasonal human coronaviruses - OC43, HKU1, 229E, and NL63 – only cause mild respiratory infections, three highly pathogenic coronaviruses that emerged in the last two decades revealed the pandemic potential of this family of viruses. It is known that severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1) and Middle East respiratory syndrome coronavirus (MERS-CoV) can cause acute respiratory distress syndrome and even death. The fatality rates of these viruses ranged from 10 to 40%. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which caused the current COVID-19 pandemic, has a lower fatality rate but is far more transmissible than SARS-CoV-1 and MERS-CoV. So far, it has been responsible for nearly 33 million cases and 996,000 deaths worldwide. Given the severity of the impact of these viruses on human health, we must understand the mechanism behind the invasion of host cell machinery by SARS-CoV-2 and other coronaviruses during infection. Identifying host factors common to several coronaviruses could help develop therapies that can fight current and future pandemics caused by coronaviruses. CRISPR screens of infected cells reveal distinct viral entry factorsIn a recent preprint paper published on the preprint bioRxiv,* researchers from the University of California San Francisco, Gladstone Institutes, Chan Zuckerberg Biohub, and Synthego Corporation, San Francisco, discuss how they identified some host factors common to 3 coronaviruses with the help of the gene-editing tool, CRISPR. The team of researchers conducted parallel genome-wide CRISPR screens cells infected with SARS-CoV-2 and two seasonal common cold coronaviruses - OC43 and 229E. They were able to identify the distinct viral entry factors for all 3 viruses - ACE2 for SARS-CoV-2, glycosaminoglycans for OC43, and aminopeptidase N for 229E. Moreover, they also found that phosphatidylinositol phosphate biosynthesis and cholesterol homeostasis are critical host pathways that support infection by these three coronaviruses. TMEM106B, the lysosomal protein, was unique to SARS-CoV-2 infection. By pharmacological inhibition of cholesterol homeostasis and phosphatidylinositol phosphate biosynthesis, the authors were able to reduce the replication of all three coronaviruses. 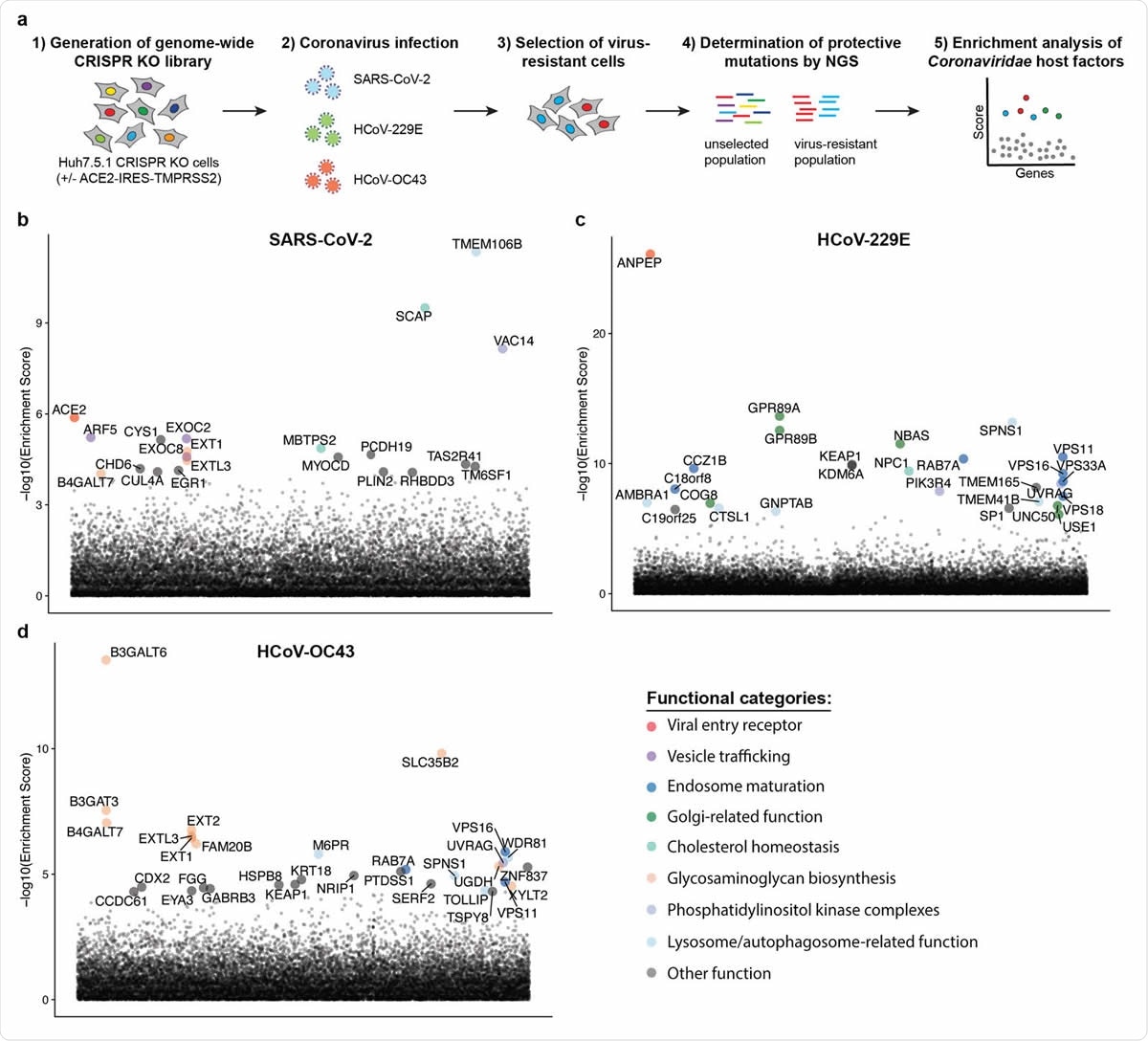 Genome-wide CRISPR KO screens in human cells identify host factors important for infection by for SARS-CoV-2, HCoV-229E and HCoV-OC43. (a) Schematic of CRISPR KO screens for the identification of coronavirus host factors. Huh7.5.1-Cas9 (with bicistronic ACE2-IRES-TMPRSS2 construct for SARS-CoV-2 and without for 229E and OC43 screen) were mutagenized using a genome-wide sgRNA library. Mutant cells were infected with each coronavirus separately and virus-resistant cells were harvested 10-14 days post infection (dpi). The abundance 347 of each sgRNA in the starting and selected population was determined by high-throughput sequencing and a gene enrichment analysis was performed. (b-d) Gene enrichment of CRISPR screens for (b) SARS-CoV-2, (c) 229E and (d) OC43 infection. Enrichment scores were determined by MaGECK analysis and genes were colored by biological function. The SARS-CoV-2 was performed once. The 229E and OC43 screens were performed twice and combined MaGECK scores are displayed. Data suggests unique entry factors but common host pathways for 3 coronavirusesThe results of the study highlight that while the three coronaviruses depend on unique entry factors, they have a common set of host pathways that help in infection. Genes linked to cholesterol homeostasis were highlighted in all screens and the network propagation. "Due to its involvement in multiple cellular processes including vesicular trafficking and autophagy, it remains to be determined whether coronaviruses hijack this pathway during entry or for the generation of double membrane vesicles required for the viral replication/transcription complexes." Consistent with the findings, 2 SARS-CoV-2 interactomes revealed viral proteins binding to SCAP, which was identified as a host factor that is critical for the replication of the SARS-CoV-2 virus. The researchers argue that given the role of SCAP in viral replication, viral proteins likely positively regulate SCAP activity and cholesterol levels. These findings provide key insights into the understanding of the life cycle of coronaviruses and important directions for the development of host-directed therapies to combat coronavirus infection. Previous studies have linked cellular cholesterol homeostasis to viral entry and membrane fusion in bunya- and hantavirus infections. This suggests a pro-viral function across different families of viruses. The team's findings were in agreement with this hypothesis. They were able to reduce infection with SARS-CoV-1 and CoV-2 spike-pseudotyped viruses by treatment with 25-hydroxycholesterol, which blocks SREBP processing and inhibits cholesterol synthesis. The results are also consistent with that of a recent drug repurposing screen that identified more than 100 compounds, including PIKfyve inhibitors, protease inhibitors, and cholesterol homeostasis modulators, that inhibited SARS-CoV-2 replication. The functional genomics data obtained by this study suggests that the observed effects of these compounds were potentially due to inhibition of crucial host factors. "Our results corroborate previously implicated host pathways, uncover new aspects of virus-host interaction, and identify targets for host-directed antiviral treatment." According to the team, the study offers critical insights into host pathways, usually hijacked by coronaviruses. They identified the phosphatidylinositol PIK3C3 kinase complex as a potential therapeutic target for SARS-CoV-2 based on the 229E and OC43 screens, highlighting the value of the parallel CRISPR screening for developing novel therapies against SARS-CoV-2 and other viruses of the Coronaviridae family. *Important NoticebioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information. Journal reference:
|
| In deadly COVID-19 lung inflammation, discover a culprit in NFkB pathway - Science Daily Posted: 29 Sep 2020 12:00 AM PDT Scientists at Boston University's National Emerging Infectious Diseases Laboratories (NEIDL) and the Center for Regenerative Medicine (CReM) joined forces to develop the most relevant research model possible for understanding how the coronavirus virus impacts the lungs, by engineering living, "breathing" human lung cells from stem cells for the task. Their efforts have borne a leap forward in our understanding of how COVID-19 infections trigger deadly levels of lung inflammation. Their discovery of a pathway that sets the lungs ablaze with inflammation has launched a search for new therapeutics that could block this process before it can take off and turn fatal. According to their new findings, published in Cell Stem Cell, the trouble starts soon after the air sacs in the lungs are infected with SARS-CoV-2, when the virus activates one of the body's biological pathways known as NFkB (the k is pronounced "kappa"). As that's happening, the virus also suppresses the lungs' ability to call in the help of the immune system to fight off the viral invaders. When the signal for help finally goes out -- several days after infection has taken hold -- an army of immune cells swarms into lung tissue heavily laden with infected, dead, and dying cells and with unchecked levels of inflammation triggered by the early activation of NFkB. The incoming immune cells, by attempting to destroy every infected cell in their path, add more fuel to the fire. Every infected cell killed by the virus or by immune cells trying to thwart viral spread tips the scales of inflammation closer to sending the lungs and other organs into total failure. The discovery of NFkB's role in this deadly cascade makes it a promising target for new therapeutics that could tamp down its activity early on after infection with the novel coronavirus. A new drug could help reduce inflammation before it gets out of control, and give the body critical time to recruit help from immune cells before conditions have deteriorated too far."We've learned [NFkB] is the primary pathway that drives inflammation [in COVID-19 patients]," says one of the study's corresponding authors, Darrell Kotton, who is a director of the CReM and a pulmonologist at BU's teaching hospital, Boston Medical Center (BMC). "Now the challenge is to find effective therapeutics that work in patients who are developing acute respiratory distress syndrome [ARDS]."Kotton says the revelation about NFkB's role in severe coronavirus infections is important because the data was gathered directly from observing human lung cells infected with live SARS-CoV-2 virus. That's different from the vast majority of coronavirus research written about to date, which has been based on infecting much more commonly available types of cells for research: kidney cells derived from African green monkeys. Those kidney cells are easily grown and maintained in culture dishes, but don't accurately represent human lung cells. "You learn a lot more about how human beings respond to the virus and how drugs might work in them when you infect human lung cells, not kidney cells from monkeys," Kotton says. For study co-corresponding author Elke Mühlberger, a virologist at the NEIDL who typically works with some of the world's most lethal viruses, like Ebola and Marburg, it was remarkable to witness the effect SARS-CoV-2 virus has on human lung cells. "It was scary to see how much damage the virus does to these cells," Mühlberger says. "It disrupts the [membrane surrounding the cell nucleus], and causes significant changes to the cell's organelles," which are the internal parts of a cell that carry out essential functions. "The cells really suffer," she says, and not even Ebola or Marburg viruses have as much impact on the cell's internal organelles as the novel coronavirus does, she adds. "I don't think the senior members of our research team and I have ever experienced anything like this in our careers," Kotton says. He and co-corresponding author Andrew Wilson, also a CReM scientist and a pulmonologist at BMC, have avoided stepping foot in their own labs in the CReM in order to protect their colleagues from the coronavirus exposure risks they endure inside BMC's intensive care unit. At BMC, Kotton and Wilson frequently saw patients infected with the coronavirus who, despite having mild cold-like symptoms and feeling pretty healthy for a week or so, would suddenly crash, needing to be intubated and ventilated. "We saw this process taking place right in front of our eyes. It was so evident to us that we were trying to keep patients alive after the damage to their lungs had already happened," Kotton says. That fueled their interest in looking at what's happening inside the lungs' air sac cells at the onset of coronavirus infection. "We had to get a glance at what the cells are doing when the disease first takes off, because after that, it's probably too late to stop the process except to help keep patients alive with a ventilator," Kotton says. "The first day a lung cell gets infected, what is the cell telling us? We hypothesized that time frame might be a much more effective window to intervene." Peering into that window, they identified NFkB as the primary culprit. To make that discovery, Adam Hume, a co-lead author of the study and a senior research scientist in Mühlberger's lab, performed the SARS-CoV-2 infections on sophisticated lung models created by stem cell researchers in Kotton and Wilson's CReM labs -- Jessie Huang, Rhiannon Werder, and Kristy Abo, also co-lead study authors. Their models of human lung tissue -- three-dimensional structures of lung cells, called "lung organoids" -- are grown from human stem cells. The CReM's organoids have been used by researchers at BU and with collaborators elsewhere to study a range of chronic and acute lung diseases. For coronavirus research, CReM scientists leveraged their organoid expertise to grow lung air sac cells, the type of cell that lines the inside of lungs. Air sac cells are usually difficult to grow and maintain in traditional culture and difficult to extract directly from patients for research purposes. That's why many labs rely on the use of more readily available cell types, like kidney cells from monkeys. "Our organoids, developed by our CReM faculty, are engineered from stem cells -- they're not identical to the living, breathing cells inside our bodies, but they are the closest thing to it," Kotton says. The CReM team then placed the human air sac cells into an experimental model they had previously developed to study the effects of smoking cigarettes. The cells are plated on a mesh membrane; on one side they are exposed to air, just like air sac cells experience in the lungs when we breathe. On the other side of the membrane, the cells are fed by a liquid concoction that mimics the nutrients and growth factors supplied by lungs' blood vessel network. From there, the NEIDL team stepped in to infect the lung model. Hume added droplets of live coronavirus on top of the lung cells, infecting them from the air side of the membrane, similar to the way the virus infects cells lining the inside of the lungs when air containing the virus is breathed into the body. He and Mühlberger have run these experiments inside one of the NEIDL's Biosafety Level 4 (BSL-4) laboratories, the highest possible level of biosafety containment used for infectious agents that pose especially high risk to humans. Based on the experiments implicating NKfB's role in severe coronavirus cases, the CReM and NEIDL researchers are now collaborating with researchers at BU and beyond who have libraries of drugs and novel chemical compounds. Together, the collaborators plan to screen for potential therapeutics that could block the train of inflammation from leaving the station. Using the CReM lung model at the NEIDL, the researchers have confirmed that existing drugs remdesivir and camostat are effective in combating the virus, though neither is a perfect fix for controlling the inflammation unleashed by NFkB. Remdesivir, a broad-use antiviral, has already been used clinically in coronavirus patients. Camostat, an antiviral and cancer drug sometimes used to treat pancreatitis, has previously been tested in a type of cell found higher up in the lungs, in the airway, and found to be effective. With the BU team's experiments confirming it also works to treat coronavirus infection in the lungs' air sac cells, Kotton says camostat is a good candidate for clinical trials. "It's been wonderful to finally be on the offense, rather than on the defense, like we were early on in April and May," Wilson says. At BMC, he recalls during the springtime surge of coronavirus cases, "There was a period of time where overhead announcements, calling code teams to assist with a patient doing poorly, would go off at least once an hour. It was really, really intense." As patients struggled to survive, Wilson says he and other clinicians felt like there was relatively little they could offer in terms of specific therapies for the coronavirus. "That was so hard -- you so badly want to do something to help someone get better," he says. Now, untangling the workings of the virus through research, he feels there has been an important change of mindset. "How can we attack this virus? The cells we're using in these experiments are the cell type most prominently affected in sick patients, the patients we're caring for who have developed ARDS," Wilson says. "It's really important to do research on the right type of cell. It tells us how the virus is working and also what parts of the body's normal immune mechanisms aren't working like they're supposed to." |
| You are subscribed to email updates from "what is respiratory distress,what is sars,what is sars virus" - Google News. To stop receiving these emails, you may unsubscribe now. | Email delivery powered by Google |
| Google, 1600 Amphitheatre Parkway, Mountain View, CA 94043, United States | |
.jpg)
Comments
Post a Comment